Ammm:Flexible Docking with GOLD and GLIDE
Introduction
Many biological processes involve binding of a protein to some target model:
- Signaling mechanism between cells
- Mechanical operation (e.g. muscle contraction)
- Mediation of a catalytic event
- ...
Drugs can work as competitive inhibitors by binding to proteins more strongly than their natural partners.
Laboratory techniques for drug discovery are very time-consuming and expensive, therefore, there is a great deal of interest in developing accurate and efficient computational techniques.
Docking can be defined as the simulation of molecular recognition and the prediction of structure of receptor-ligand complexes where:
- Receptor is a protein (or protein oligomer)
- Ligand is either a small molecule or another protein
Docking programs generally have two major components:
- Search algorithm
- Scoring function
GOLD
GOLD is a flexible protein-ligand docking program developed by Jones et al. in 1995 [1] [2].
Genetic algorithm
A genetic algorithm is a search technique used in computing to find exact or approximate solutions to optimization and search problems. It is implemented as a program that mimics evolution by manipulating a population of individuals represented by data structures called chromosomes.
Each structure encoded in a chromosome is a possible docking solution, and it:
- Represents the conformation of the ligand and its position (rotation, translation) in the docking site, and
- Has a fitness score assigned to it (given by scoring function); this score is used to assess how good a solution this individual is.
In GOLD, a steady-state operator-based genetic algorithm with no duplicates is used.
- In a steady-state genetic algorithm, only a single population of individuals is maintained at any given time, and newly created individuals are added to the population using a replacement operator, which selects individuals from the parent generation to be removed.
The genetic algorithm implemented also makes use of the island model, which is a practical and efficient way of parallelizing the algorithm.
- n sub-populations are created and arranged in a ring, such that each sub-population has only two neighbors
- Genetic operators are applied to individual sub-populations in turn.
- When the migration operator is selected, a child is created as an identical copy of a parent, and "migrates" to a neighboring sub-population.
The algorithm in GOLD is the following:
- 1. Choose a set of reproduction operators, each with a weight assigned to it.
- 2. Create an initial random population. Calculate the fitness of its members.
- 3. Choose an operator to apply using roulette wheel selection based on operator weights.
- 4. Choose parents required by operator by using roulette wheel selection again, this time based on individuals' fitness.
- 5. Apply operator to produce child chromosome. Evaluate new individual's fitness.
- 6. If not redundant of a member in previous generation, replace least fit member with new individual.
- 7. If 100,000 operators have been applied, STOP. Else, GOTO step 3.
Initializing the protein and the ligand
Protein
- Water molecules and ions are removed.
- Hydrogens are added at appropriate geometry.
- User indicates approximate binding site by supplying values for (ORIGIN, RADIUS). From these values, interaction region is computed and cavities are detected.
- Hydrogen bonding sites are calculated at the interface.
Ligand
- Hydrogen atoms are added.
- Hydrogen bond donors and acceptors are identified.
- Ligand is fully minimized using SYBYL's MAXIMIN2 module.
- A random translation, rotation and conformation are chosen for the ligand so as not to start with a bias.
Chromosome representation
Two binary strings are used to specify conformation for:
- protein
- ligand
Each byte encodes the angle of rotation about each bond, such that , where the angle can increase or decrease by a step size of .
Two integer strings are used to encode mappings in hydrogen bonds determined by least squares fitting:
- mappings from acceptors in ligand to donors in protein
- mappings from donors in ligand to acceptors in protein
The value is null if no bond is formed.
Fitness function
The fitness of a given solution is evaluated in six steps:
- 1. Conformation of ligand and protein's active site are generated.
- 2. Ligand is placed in active site using least squares fitting.
- 3. Hydrogen bonding energy is calculated for complex.
- 4. Pairwise energy is obtained for steric energy of interaction between protein and ligand.
- The 8-4 potential is chosen because it is softer than the 12-6, and it allows for closer contacts with the protein to be formed more easily.
- 5. Internal energy is calculated for ligand using molecular mechanics terms.
- This term is the sum of the ligand's steric and torsional energies.
- The steric energy is calculated by:
- The Tripos forcefield torsional potential used is of the form:
- Where i,j,k and l are four consecutively bonded atoms, is the torsional angle, n is the periodicity, and V is the barrier to rotation.
- This term is the sum of the ligand's steric and torsional energies.
- 6. All energy terms are summed up to get final fitness score.
Genetic operators
Three different types of operators can be applied in GOLD's genetic algorithm:
1. Crossover
- Requires two parents
- Produces two children
- Performs one-point crossover on binary strings
- Performs two-point crossover on integer strings
2. Mutation
- Requires one parent
- Produces one child
- Position randomly chosen on string, and value at that position replaced with different new value from a set of acceptable values. If this value occurs elsewhere on the string, replace it with original value of mutated position.
3. Migration
- Requires one parent
- Produces one child
- Child is created as an identical copy of a parent, and "migrates" to a neighboring sub-population.
GLIDE
GLIDE is a flexible protein-ligand docking program developed by Friesner et al. in 2004 [3] [4] [5].
Docking methodology
The GLIDE docking "funnel" showing the docking hierarchy:
GLIDE applies a series of hierarchical filters to search for possible locations of the ligand within the receptor's active site.
- The shape and properties of the receptor are mapped onto the grid during the pre-processing phase.
- The ligand's pose is the set of values for its
- position
- orientation
- core conformation
- rotamer group conformations
Protein preparation
Preparation is done in a series of five steps:
- 1. Hydrogens are added to the co-crystallized ligand and bonds are defined.
- 2. Residues that do not participate in salt bridges and are more than a specified distance from the nearest ligand atom are neutralized (pprep script). The tautomeric state for His residues (assumed neutral) is also set by considering potential metal-ligation and hydrogen-bonding interactions.
- 3. Post-processing of the receptor is performed, based on the assumption that the actions taken in the pre-processing step are not always correct, e.g. wrong charge assignment.
- 4. Structural waters are then added if necessary. Although omitting them can be useful because it allows ligands to be found that are capable of displacing them, it can also encourage false positives due to an overly "generous" active site.
- 5. Hydrogens are added to the protein. A final minimization is applied.
The preparation of the protein and ligand were found to greatly affect the quality of the docking.
Conformation generation
GLIDE starts out by performing an exhaustive conformational search.
It follows it up by a rapid elimination of conformations that it finds to be unsuitable for binding to the receptor, such as conformations that have long-range internal hydrogen bonds, by:
- evaluating the torsional energy of the various minima using a truncated OPLS-AA molecular mechanics potential function, and
- imposing a cutoff to the allowed value of the total conformational energy compared to the lowest-energy state.
Each ligand is divided into its:
- core region
- rotamer groups, each of which is attached to the core by a rotatable bond, but does not have rotatable inner bonds
The core plus all of its possible rotamer group conformations are docked as a single object. Therefore, the effective number of conformations being docked can add up to thousands or tens of thousands in the case of molecules with several rotatable bonds.
Initial screening of ligand poses
For each ligand core conformation, an exhaustive search of all possible translations and orientations is performed over the receptor's active site.
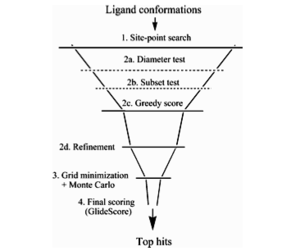
1. Selection of site points on equally spaced 2 Å grid that covers the active site.
2a. Examination of placement of atoms that lie within specified distance of ligand diameter axis. If there are too many clashes with the receptor, the orientation is skipped.
2b. The ligand is rotated about the ligand diameter axis, and the subset of atoms capable of forming hydrogen bonds with the receptor is scored.
2c. If this score is greater than a certain threshold, all interactions with the receptor are scored.
2d. The ligand as a whole is allowed to move rigidly by 1 Å in the Cartesian directions.
3. Energy minimization using a molecular mechanics scoring function: the top n poses (usually 400) are minimized on the receptor's pre-computed OPLS-AA van der Waals and electrostatics grids.
The interaction of each ligand atom with these fields is evaluated using trilinear interpolation formulae over a cube. This step can be performed for rigid or flexible docking; in the former case, only translations and rotations are taken, whereas in the latter case, torsional motion about the core and rotatable ends is also taken into account.
Three or six lowest-energy poses obtained are then subjected to Monte Carlo minimization.
4. Finally, these minimized poses are re-scored using a model energy score, "Emodel" which combines:
- energy-grid score
- binding affinity predicted by GlideScore
- internal strain energy for the model potential used to direct the conformational search algorithm (for flexible docking)
Scoring function
GLIDE has two forms of scoring functions: GlideScore Standard-Precision (SP) and GlideScore Extra-Precision (XP).
The functions have similar terms, but are formulated with different objectives in mind:
- GlideScore SP offers a "softer" docking, minimizing false negatives, whereas
- GlideScore XP applies harsher penalties for poses that violate physico-chemical principles, limiting the number of false positives.
GlideScore SP
This scoring function is mostly derived from the empirically-based scoring scheme used in ChemScore [6].
It first assigns general atom types to all ligand atoms and receptor atoms in contact with the ligand:
- Lipophilic: Chlorine, bromine and iodine atoms which are not ions; sulphurs which are not acceptor or polar types; carbons which are not polar.
- H-bond donor: Nitrogens with hydrogen attached; hydrogens attached to oxygen or nitrogen.
- H-bond donor/acceptor: Oxygens attached to hydrogen atoms; special case of imine nitrogen (i.e. C=NH nitrogen).
- H-bond acceptor: Oxygens not attached to hydrogen; nitrogens with no hydrogens attached and one or two connections; halogens which are ions; sulphurs with only one connection.
- Polar (non H-bonding): Nitrogens with no hydrogens attached and more than two connections; phosphorus; sulphurs attached to one or more polar atoms (including H-bonding atoms and not including polar carbon atoms or fluorine atoms); carbons attached to two or more polar atoms (including H-bonding atoms and not including polar carbon atoms or fluorine atoms); carbons in nitriles or carbonyls; N atoms with no hydrogens and four connections; fluorine atoms.
- Metal: Metal atoms.
The scoring function is of the form:
The f, g and h functions give a full scores (1.00) for distances and angles that lie within normal limits, and a partial score (1.00-0.00) to distances and angles that lie outside that range, but inside larger threshold values. E.g. in the hydrogen bonding term, if the H...X hydrogen bond distance is within 0.25 Å of a nominal value of 1.85 Å then is 1.00. If, however, the hydrogen bond distance lies between 2.10 and 2.50 Å, then it tails off to zero in a linear fashion.
The seven different terms are the following:
1. Lipophilic-lipophilic: calculated for all lipophilic ligand atoms and all lipophilic receptor atoms.
2-4. Hydrogen-bonding: split into three to improve scoring; the first of these contributions is the most stabilizing, and the last is the least important. It is calculated for all complementary possibilities of hydrogen bonds between ligand and receptor atoms.
5. Metal ions: calculated for all acceptor and acceptor/donor atoms in the ligand and any metal atoms in the receptor. The function f(r) is a simple contact term and its functional form is illustrated in:
6. Rotatable bonds: identifies "frozen" rotatable bonds, i.e. where atoms on both sides of the bond are in contact with the receptor.
- where is the number of frozen bonds, the summation is over frozen bonds, and and are the percentages of non-lipophilic heavy atoms on either side of the rotatable bond.
7. Polar-phob: rewards instances in which polar but non-hydrogen bonding atom is found in hydrophobic region.
8. Coulombic interaction energies.
9. Van der Waals interaction energies.
10. Solvation model: explicit waters are docked into the binding site for each energetically competitive ligand pose, and empirical scoring used to measure exposure of various groups to explicit waters.
GlideScore XP
GlideScore XP is a more recent version of the GLIDE scoring function. It has two novel features:
- application of large desolvation penalties to both protein and ligand polar and charged groups in appropriate cases, and
- identification of specific structural motifs that provide exceptionally large contributions to enhance binding affinity.
It is written as:
Where the binding energy is:
And a penalty is incurred:
New terms contributing to binding include:
- Hydrophobic enclosure (): assigns scores to lipophillic ligand atoms based on summation over a pair function, each term of which depends on interatomic distance between ligand atom and neaighboring lipophillic receptor atom. The displacement of water molecules from areas with many proximal lipophillic protein atoms results in lower free energy than areas with fewer such atoms.
- Special Neutral-Neutral Hydrogen Bond Motifs ()
- Special Charged-Charged Hydrogen Bond Motifs ()
- Pi Stacking and Pi-Cation Interactions ()
An important addition is the set of terms that penalize binding:
- Water Scoring: Rapid Docking of Explicit Waters ()
- Contact Penalties (): most difficult component of empirical scoring function. The problem arises from the fact that the ligand has to adjust to fit in an imperfect and rigid cavity, and so, might adopt a higher-energy, non-ideal conformation. Furthermore, even native ligand geometries can exhibit high strain energies. Therefore, this term is rather lenient, penalizing only severe cases of bad internal clashes.
Comparison
Results from comparative study by Kellenberger et al. [7].
Software Links
Further Reading







References
- ↑ Jones, G., Willett, P., Glen, R.C., Leach, A.R. and Taylor, R. Development and Validation of a Genetic Algorithm for Flexible Docking, 1995. J. Mol. Biol. 267: p. 727-748.
- ↑ Jones, G., Willett, P. and Glen, R.C. A genetic algorithm for flexible molecular overlay and pharmacophore elucidation, 1995. Journal of Computer-Aided Molecular Design. 9: p. 532-549.
- ↑ Friesner, R.A. et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy, 2004. J. Med. Chem. 47: p. 1739-1749.
- ↑ Halgren, T.A. et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening , 2004. J. Med. Chem. 47: p. 1750-1759.
- ↑ Friesner, R.A. et al. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein-Ligand Complexes, 2006. J. Med. Chem. 49: p. 6177-6196.
- ↑ Elridge, M.D. et al. Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes, 1997. Journal of Computer-Aided Molecular Design. 11: p. 425-445.
- ↑ Kellenberger, E., Rodrigo, J., Muller, P. and Rognan, D. Comparative Evaluation of Eight Docking Tools for Docking and Virtual Screening Accuracy, 2004. PROTEINS: Structure, Function and Bioinformatics. 57: p. 225-242.






![{\displaystyle E_{ijkl}={\frac {1}{2}}V_{ijkl}[1+{\frac {n_{ijkl}}{|n_{ijkl}|}}cos(|n_{ijkl}|\cdot \omega _{ijkl})]}](https://wikimedia.org/api/rest_v1/media/math/render/svg/21a02812156b4183643a197b18077300e84e2545)
























